
WEHI Home Page
About WEHI
News
Research
Education
Intranet
(Staff Only)
Search
Contact WEHI
© Copyright 2002
Walter & Eliza Hall Institute
Contact the Webmaster

About limmaGUIlimmaGUI is a Graphical User Interface for Gordon Smyth's limma package (Linear Models for MicroArray data). It uses state-of-the-art statistical techniques to normalize microarray data, perform diagnostic plots and to find differentially expressed genes in complex experimental designs. limma and limmaGUI are both R packages. The limma package offers R users a command-line interface. The limmaGUI package, while not as powerful as limma to the expert user, offers a simple point-and-click interface to many of the commonly-used limma functions. In order to use limmaGUI, you need to have R 1.9.0 or later, Tcl/Tk 8.3 or later (ActiveTcl for Windows or X11 Tcl/Tk for MacOSX or an appropriate Linux Tcl/Tk installed either from source or by a package manager e.g. RPM), including the Tktable and BWidget Tcl/Tk extensions, both of which come with ActiveTcl. You also need the limma, limmaGUI, sma, tkrplot, R2HTML and xtable R packages. The limma package is available on Bioconductor (Click on Release 1.3 packages), the limmaGUI package is available here, and the other packages are available from CRAN. limmaGUI has been succesfully tested on Windows 2000, Windows XP, and Red Hat Linux, and on Mac OSX with X11. Some screenshots from limmaGUIThe Weaver data set... 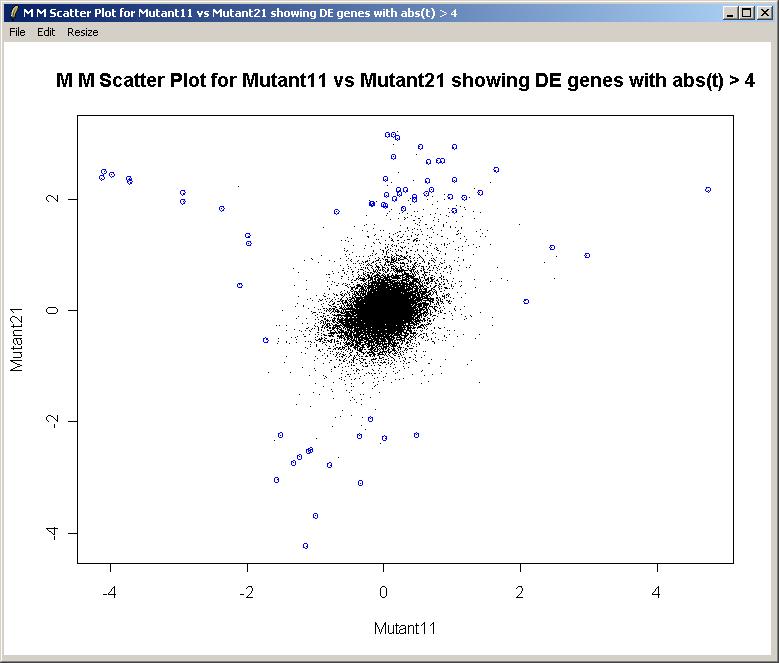 Differentially Expressed Genes from the Swirl Data Set...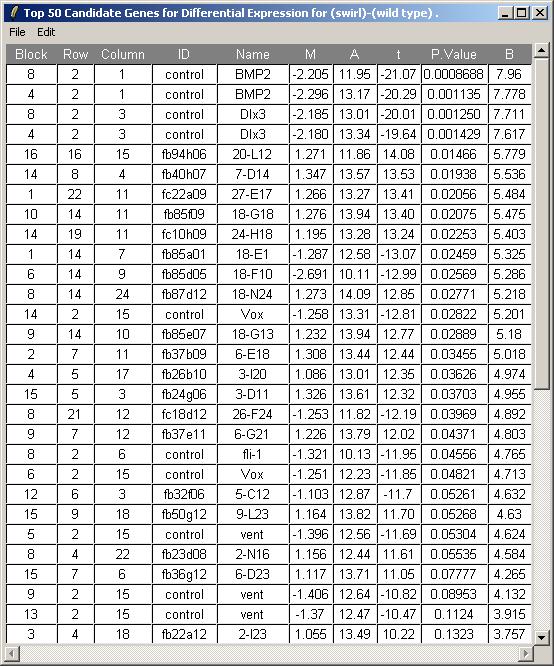 limmaGUI Home
Comments/Questions? Contact wettenhall@wehi.edu.au. |